2. 大连理工大学, 辽宁 大连 116024
2. Dalian University of Technology, Dalian Liaoning 116024, China
基因是遗传的主要功能单位,它们所包含的核酸碱基的特定序列能够编码生物体功能所需的大部分蛋白质。作为基因的载体,2种类型的核酸:核糖核酸(Ribonucleic Acid,RNA)或脱氧核糖核酸(Deoxyribonucleic Acid,DNA)受到广泛关注,尤其是基于核酸的体外分子检测技术,成为强大的生物学研究工具[1-2]。基于核酸的PCR(Polymerase Chain Reaction)诊断比传统方法(基于酶或者抗体的分析)具有更多的优势:更高的修改灵活性、更高的检测灵敏度以及更快、更早的获得检测结果[3],在人类疾病控制(如艾滋病[4]、疟疾[5]和丙型肝炎[6]等)、农作物病害控制[7]、畜牧业疫病防控[8]和食品安全领域[9]均发挥着极其重要的作用。PCR诊断包括多个关键环节:核酸的提取,聚合酶链反应(PCR)及结果读出[10]。从生物体中提取核酸,需要将核酸从含有蛋白质、多糖和脂肪等多种生物大分子的复杂体系中进行分离、纯化,是PCR、测序等生物学分析的先决条件[11]。核酸提取的质量,会显著影响后续分析的准确性和灵敏度。生物样本中残留的抑制性成分(血红素、球蛋白等成分)及分离、纯化过程中残留的有机物和盐分,是影响PCR灵敏性和分析稳定性的重要因素[12-14]。
传统的核酸的分离方法是基于有机溶剂萃取的液相分离,如Chomczynski’s法[15]、酚氯抽提方法[16-17]、溴化乙锭-氯化钙梯度离心法等[18]。液相分离方法虽然有效,但存在着很大的局限性:所需样品量大、分离时间长、易受污染、降解率高[19]。另外,液相分离方法中有机溶剂的残留显著增加了PCR抑制剂的残留风险。为了提高提取核酸样品的纯度与浓度,核酸的提取方法也在不断更新,目前固相萃取核酸的方法受到更多学者的青睐。固相萃取核酸的方法主要是采用一些对核酸有结合作用的固相材料,在一定条件下结合核酸然后又在适当条件下脱附,实现核酸的分离。与液相萃取方法相比,固相萃取方法减少了有机溶剂的使用,操作相对简便,分离时间短,样品在实验过程中不易污染与降解,提取的核酸无论是浓度还是纯度都相对较高[20-23]。本论文通过综述具有核酸结合能力的固相材料及其在核酸固相分离过程中的研究进展,探讨核酸固相分离在病原PCR诊断应用中的优势和局限,预测其发展趋势。
1 具有核酸结合能力的固相材料及其作用机制固相核酸分离主要是通过对核酸有吸附作用的固体颗粒在一定条件下结合核酸而实现的分离。目前,报道的具有核酸结合功能的固相材料主要包括:二氧化硅及其衍生物[24-30]、金属及其衍生物[31-35]、云母[36]、合成树脂[37-38]、纤维素[39]和氧化石墨烯[40-41]等,通过静电作用、共价键结合等机制结合、分离核酸。
1.1 二氧化硅及其衍生物二氧化硅纳米颗粒作为无机纳米粒子,具有化学性质稳定、价格低廉和比表面积大的特点,其可以均匀地分散在裂解液体系中,能够更有效地结合核酸,因此二氧化硅及其衍生物被广泛用于核酸固相分离[28, 42-43]。二氧化硅吸附核酸的方法主要分为:盐桥法、直接静电法和配位法[44]。盐桥法中二氧化硅颗粒吸附核酸的驱动力包括静电屏蔽作用、脱水作用以及分子间氢键作用。在高浓度盐中颗粒与核酸之间的静电斥力会被屏蔽;当盐浓度达到1个临界值时,颗粒与核酸足够近的情况下可以通过分子间氢键的作用使得核酸吸附在二氧化硅颗粒表面;在胍盐存在的情况下,颗粒和核酸还可以通过脱水作用结合在一起[44]。另外,二氧化硅纳米颗粒表面也可以包覆聚乙烯亚胺(Polyethyleneimine,PEI)[45]、壳聚糖[46]等聚阳离子电解质,使颗粒在特定的裂解条件下呈正电荷,通过直接静电作用与带负电荷的核酸相结合。Jiang等[47]采用双羧基偶联剂与二氧化硅纳米颗粒偶连,使得其表面有双羧基能够鳌合Fe3+,进而通过Fe3+与磷酸根的配位作用来结合核酸。
1.2 金属及其衍生物金纳米粒子(Au-Nanoparticles,Au-NPS)作为金属材料,具备与四种脱氧核苷结合的良好的亲和力,被广泛用来结合核酸分子[31, 48-49]。由于双链DNA(Double Stranded DNA,ds-DNA)的结构复杂[50],且具有比单链DNA(Single Strand DNA,ss-DNA)更高的电荷密度,不利于与表面带负电荷的Au-NPS结合,因此Au-NPS主要用来结合ss-DNA,且该结合仅在高温下完成。另外有研究发现Au-NPS与ss-DNA的结合随着DNA链的长度增加而减弱,且粒子的粒径大小在二者的结合力强度方面起着非常关键的作用[51]。
另外,学者们对银纳米粒子(Ag-Nanoparticles,Ag-NPS)结合特定的DNA序列的检测技术也进行了广泛研究[52-54]。Basu等[53]发现银纳米粒子(Ag-NPS)与DNA中的核酸碱基之间存在非常紧密的亲和力,根据Gu等[55]的报道这是因为Ag-NPS可以与DNA分子的碱基鸟嘌呤(G)和腺嘌呤(A)的N7原子以及碱基胞嘧啶(C)和胸腺嘧啶(T)的N3原子结合。Yang等[54]采用原子力显微镜观察发现Ag-NPS与ss-DNA可以实现有效的结合,而与ds-DNA之间却是彼此独立的随机排列。
除金、银纳米颗粒外,四氧化三铁纳米颗粒,作为一种磁性纳米颗粒,可以通过共价键结合核酸[24],只是鲜少用四氧化三铁纳米颗粒直接进行核酸分离,因为四氧化三铁纳米颗粒在结合核酸后的超顺磁性减弱或者消失[56-57],则不能发挥磁力分离的高通量和自动化优势。因此,多是在其表面负载二氧化硅及其衍生物、进行功能基团修饰[58-59],通过静电作用等结合核酸,用于快速分离。
1.3 其他固相材料云母[36]作为一种铝硅酸盐,可以通过共价键来结合核酸,或者对其进行化学修饰使其带正电荷,通过静电作用结合带负电的DNA分子。同样,合成树脂,如DEAE-Sephaeose[60]往往也是带正电荷的,可以通过静电作用与核酸结合。另外,DNA与单壁碳纳米管[61]、氧化石墨烯[41]可以通过π堆积作用结合。Zhao等[62]通过模拟研究得到的结果显示C60可以在水溶液中与核苷酸紧密结合,验证了核酸与碳材料物质间的结合。当然还有一些其他的材料可以用于核酸分离,如纤维素[39]可以与核酸共价连接。
2 核酸固相分离及其在病原PCR诊断中的应用固相核酸分离方法主要是基于一些对核酸有吸附作用的聚合物材料,出于提高吸附量的目的,具有高比表面积和多孔结构的纳米颗粒材料备受青睐。随着磁性材料的发展,1997年,Hawkins等[63]提出了核酸磁力分离——基于磁性材料快速分离核酸,并迅速发展,成为如今主要的核酸固相分离手段。下面,我们将从核酸磁力分离和非磁力分离综述近年来核酸固相分离的进展及其在分子检测中的应用。
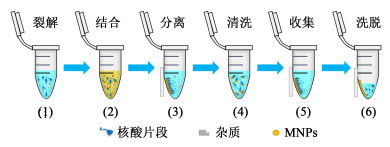
2.1 磁力分离的原理磁力分离是基于磁性纳米颗粒的一种固相分离手段,其中磁性纳米颗粒是指纳米级的粒子,一般为核壳型结构,由Fe、Co、Ni及其氧化物等组成内核,核外层则由高分子聚合物组成壳层,具有超顺磁性、比表面积大、无毒和粒径可控等优点[64-69]。这种基于磁性颗粒的核酸磁力分离,通过在一定溶液条件下,对核酸进行选择性吸附,吸附核酸的磁性纳米颗粒在外加磁场(如磁铁)时可控移动,从而从含有蛋白质、多糖等杂质的体系中分离出来,再通过洗涤、脱附,实现核酸的提取,全过程通常可在10 min内完成[70-71]。分离过程示意图如图 1所示。磁力分离过程因为避免了离心、过滤等繁琐操作,有效提高了分离速度,可以实现自动化操作,现已有基于磁力分离原理的核酸自动提取仪实现了商业化,通过自动化操作不仅极大限度地节约了人力资源,同时也避免了诸多手动操作中的误差[70]。
|
| 图 1 磁力分离过程示意图 Fig.1 Schematic diagram of magnetic separation |
| |
由于二氧化硅具备良好的结合核酸的能力,二氧化硅包覆磁流体形成的Fe3O4@SiO2微球颗粒是最常用的核酸磁力分离纳米材料。将二氧化硅涂覆到Fe3O4表面,主要方法有微乳液法[72]和Stöber法(原硅酸四乙酯的碱水解法)[73-76]。Stöber法因为不存在表面活性剂,合成后的颗粒保留了快速的磁响应能力,同时表面二氧化硅层可以防止颗粒之间的聚集,分散性好,更适合生物应用[77-78]。然而Stöber法中的多步修饰程序使制备过程既耗时又费力,Ma等[79]因此开发了一种溶剂热法,可以更简单、低成本地制备形状均一、单分散性的Fe3O4@SiO2纳米颗粒,将其用于大肠杆菌的核酸分离,可以获得高品质无蛋白质污染的核酸用于PCR扩增。Fan等[80]则将溶剂热法改进,首先使用四氯化硅作为交联剂修饰Fe3O4颗粒表面, 继而实现颗粒表面硅层的快速可控生长,所得纳米颗粒具有优异的耐酸性和稳定、致密的硅涂层,用于质粒DNA分离具有比商业化磁珠更高的产率。
随着二氧化硅负载技术的不断发展,越来越多的磁性纳米粒子被开发出来,得到了普遍优于市售磁性纳米颗粒的应用效果。Dao等[81]制备了二氧化硅包覆的磁性纳米颗粒,用于提取乙型肝炎病毒(Hepatitis virus type B,HBV)和巴尔病毒(Epstein-Barr Virus,EBV)的基因组DNA,结果显示,该磁性颗粒提取的DNA在PCR扩增中产物条带在紫外光下清晰、明亮,具有比市售磁性颗粒更好的DNA纯化效率。Xu等[82]制备的二氧化硅包覆的具有理想磁化强度的磁性复合微球可作为有效的质粒DNA富集载体,相较于市售SM1-015B微球有很大优势, 能够得到更大的提取效率与更优的质量。Deng等[83]制备了以Fe3O4@SiO2为核、垂直排列的介孔二氧化硅微球,具有均匀的孔径(2.3 nm),借助其较大的孔径和优异的磁性,能够实现微囊藻毒素的快速高效(分离效率>95%)分离。Liu等[84]合成了Fe3O4@SiO2纳米颗粒用于DNA定量分析,发现与市售微型Dynabeads(R)相比,该纳米颗粒具有更好的灵敏度来定量DNA,检测限能够达到4 000碱基对(人类DNA片段)。
二氧化硅广泛应用于包覆Fe3O4制备磁性纳米颗粒,不仅可以有效防止磁性纳米颗粒聚集,同时也方便引入其他活性基团[78]。He等[85]采用氨基修饰的二氧化硅包覆的磁性颗粒分离质粒DNA,发现从1 mL制备的细菌裂解液中可分离到约55 μg的pEGFP-N3质粒DNA,与传统的酚氯抽提方法(同样条件分离到约55 μg的pEGFP-N3质粒DNA)相比有很大的优势,且A260/A280的比值可以达到1.9,说明提取的DNA纯度很高,可以忽略蛋白质的污染。Wu等[59]采用羧基修饰的二氧化硅包覆的磁性纳米颗粒分离大肠杆菌样品中的DNA,发现提取的DNA平均浓度为16 mg/L,其A260/A280的比值为1.75(提取质量较好),且电泳结果显示大肠杆菌分子量按照预期分布,每个波段的亮度均匀,说明羧基修饰的磁性纳米粒子用于核酸提取的有效性。Pham等[86]采用二氧化硅包覆的磁性颗粒与氧化石墨烯组成的新型磁性材料,用于RNA的提取,发现得到RNA的数量是传统的酚氯抽提方法的1.7倍。Chen等[67]以戊二醛为交联剂,通过共价键将血红蛋白固定在氨基功能化的Fe3O4@SiO2磁性纳米粒子表面,得到了血红蛋白修饰的新型磁性纳米复合材料,将其应用于大肠杆菌中质粒DNA的提取,发现对质粒DNA的吸附效率可以达到93%。另外,Perçin等[87]制备了新型材料——聚甲基丙烯酸羟乙酯磁性纳米粒子,发现其对质粒DNA的总回收效率可以达到92%,且该磁性颗粒在不明显降低质粒DNA的吸附量前可被循环使用6次。
2.3 磁力分离在病原PCR诊断中的应用磁力分离作为一种高效、节省成本的分离方法,具备与核酸结合后能够在外加磁场快速移动的特点[84],可以避免传统核酸分离过程中繁琐的离心操作和诸多手动操作误差,具有下游高通量和自动化应用的前景[71]。
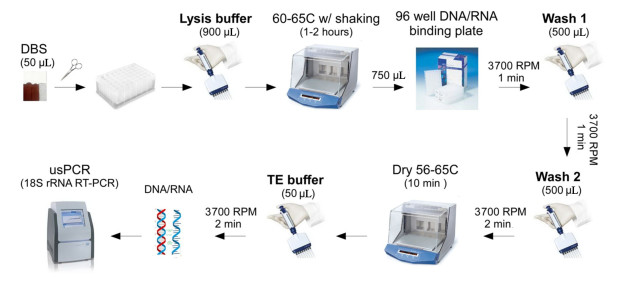
Zainabadi等[88]建立了基于二氧化硅磁性粒子高通量分离干血斑核酸(如图 2所示)的方法,全过程使用排枪在96孔板上操作,通量高,只是整体时效性尚未有显著改善(提取时间约80~120 min)。因为从复杂的生物体中提取PCR诊断所需的核酸,往往包含多个步骤:样本的裂解、核酸的分离、纯化与洗脱。样本裂解作为分离前的必需步骤,目前普遍需要耗时的温浴过程,且对动植物组织样本,低温研磨等预处理又难以避免[89],不仅不能完全发挥磁力分离的高通量优势,同时制约核酸提取的整体时效性。Li等[90]则通过优化裂解过程,排除组织样本繁琐预处理同时将裂解时间缩短至最短15 min,再结合高通量磁力分离,报道了一种在96孔板上操作的全过程高通量的核酸提取方法,继而结合可视化荧光定量PCR,建立了一种灵敏、准确的柞蚕微孢子虫快速筛查技术,为更高效、更准确的柞蚕微粒子病防疫提供了新思路(如图 3所示)。
现行非磁力固相分离多需要通过过滤、离心、移液等繁琐操作才能将结合有核酸的固相材料从裂解液中分离出来,相比于借助外加磁场即能快速移动、分离的磁性颗粒而言,往往需要更多、更密集的人力操作,效率和通量局限大。Zou等[39]利用纤维素可以结合核酸的特性,选择具有一定刚性的纤维素纸条作为核酸固相分离载体,通过在裂解液和清洗液之间转移,快速从裂解液中分离出可供PCR检测的模板,时间不超过30 s。但遗憾的是,样本传统裂解过程的耗时性与繁琐性未能得到有效解决,依旧制约着核酸提取的效率,且纤维素纸条1次只能提取1个样本,分离通量低。另外,纤维素洗脱DNA困难,需要将结合有DNA的纤维素纸条作为PCR扩增的模板,对下游PCR的选择具有局限性。
固相核酸分离,尤其是磁力分离,相比液相分离具有下游高通量和自动化应用的优势,本论文通过综述核酸结合材料的开发、结合机制、核酸固相分离以及固相分离在病原PCR诊断应用方面的进展,借以论述核酸固相分离现有的应用局限性和发展前景。
由上所述,我们不难看出,磁力分离作为高通量核酸提取极具优势,但仍普遍受限于耗时的样本裂解过程,尤其是动植物组织样本难以避免的研磨等繁琐预处理,限制了整体核酸提取过程的全过程通量及提取时效性。同时,报道指出:磁力分离中纳米颗粒解吸核酸困难、DNA提取效率普遍偏低,通过将磁性纳米粒子与DNA的复合物直接作为PCR模板可以避免这个问题[91]。但磁性颗粒对PCR扩增过程具有强抑制作用,洗脱过程不可避免[92]。此外,磁性纳米颗粒固有的团聚[57]、沉降[93]等现象,在平行处理批量样本时,很大程度上会影响分离过程的稳定性。磁力分离中,因为纳米颗粒沉降现象,核酸与磁性纳米颗粒不能充分结合而影响提取产率;纳米颗粒团聚则可能会引起清洗不完全与杂质包裹等问题,影响磁力分离的提取质量,而这些过程与磁性纳米颗粒的尺寸、磁化强度及与核酸分子间的作用力强弱等动力学因素息息相关。因此,磁力分离可能的几个发展方向是:1)开展高效生物样本裂解过程研究,系统揭示裂解动力学过程,建立高效、快速、常温裂解过程,提高核酸释放量并降低多样本平行处理时的气溶胶污染风险,为后续高效、高质量磁力分离过程提供保障;2)研发新型、高效的磁性颗粒材料,并揭示核酸“吸附-脱附”动力学过程,不断优化磁力分离过程,提高分离效率和质量,是下游病原PCR诊断灵敏度与准确性的重要保障。
非磁力核酸固相分离,除了同样需要解决样本裂解步骤的局限性,同时还需要攻克核酸分离、纯化过程中对离心、过滤和移液等的依赖。纤维素纸条30 s快速分离核酸的报道[39],让我们看到了非磁力固相分离巨大的发展前景:通过手动或自动化设备,利用机械力将核酸固相结合材料快速、直接地从复杂裂解液体系中转移出来,能获得比磁力分离更快、更稳定的分离过程,也避免了颗粒状的磁性纳米颗粒聚集、沉降带来的高通量分离过程不稳定的局限。
| [1] |
Mester P, Wagner M, Rossmanith P. Molecular enrichment for qualitative molecular pathogen detection in food[J]. Food Analytical Methods, 2018, 11(5): 1251-1256. DOI:10.1007/s12161-017-1103-z |
| [2] |
Xue T, Liang W, Li Y, et al. Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor[J]. Nature Communications, 2019, 10(1): 1-9. DOI:10.1038/s41467-018-07882-8 |
| [3] |
Yang Z, Xu L, Liu L, et al. Routine screening of blood donations at Qingdao central blood bank, China, for hepatitis B virus (HBV) DNA with a real-time, multiplex nucleic acid test for HBV, hepatitis C virus, and human immunodeficiency virus Types 1 and 2[J]. Transfusion, 2013, 53: 2538-2544. DOI:10.1111/trf.12159 |
| [4] |
Jani I V, de Schacht C. Innovations and challenges in early infant diagnosis of HIV[J]. Current Opinion in Hiv and Aids, 2019, 14(1): 55-59. DOI:10.1097/COH.0000000000000511 |
| [5] |
Bauserman M, Conroy A L, North K, et al. An overview of malaria in pregnancy[J]. Seminars in Perinatology, 2019, 43(5): 282-290. DOI:10.1053/j.semperi.2019.03.018 |
| [6] |
Dore G J, Martinello M, Alavi M, et al. Global elimination of hepatitis C virus by 2030:Why not?[J]. Nature Medicine, 2020, 26(2): 157-160. DOI:10.1038/s41591-019-0706-x |
| [7] |
Rani A, Donovan N, Mantri N. Review:The future of plant pathogen diagnostics in a nursery production system[J]. Biosensors and Bioelectronics, 2019, 145: 1-12. |
| [8] |
Patrick B N, Machuka E M, Githae D, et al. Evidence for the presence of African swine fever virus in apparently healthy pigs in South-Kivu Province of the Democratic Republic of Congo[J]. Veterinary Microbiology, 2020, 240: 340-348. |
| [9] |
Stentiford G D, Becnel J, Weiss L M, et al. Microsporidia-emergent pathogens in the global food chain[J]. Trends in Parasitology, 2016, 32(4): 336-348. DOI:10.1016/j.pt.2015.12.004 |
| [10] |
Yin J, Suo Y, Zou Z, et al. Integrated microfluidic systems with sample preparation and nucleic acid amplification[J]. Lab on a Chip, 2019, 19(17): 2769-2785. DOI:10.1039/C9LC00389D |
| [11] |
Li Z, Huang J, Yang B, et al. Miniaturized gel electrophoresis system for fast separation of nucleic acids[J]. Sensors and Actuators B:Chemical, 2018, 254: 153-158. DOI:10.1016/j.snb.2017.07.064 |
| [12] |
Sidstedt M, Radstrom P, Hedman J. PCR inhibition in QPCR, DPCR and MPS:Mechanisms and solutions[J]. Analytical and Bioanalytical Chemistry, 2020, 412(9): 2009-2023. DOI:10.1007/s00216-020-02490-2 |
| [13] |
Sidstedt M, Steffen C R, Kiesler K M, et al. The impact of common PCR inhibitors on forensic MPS analysis[J]. Forensic Science International:Genetics, 2019, 40: 182-191. DOI:10.1016/j.fsigen.2019.03.001 |
| [14] |
Matsumura S, Matsusue A, Waters B, et al. Effects of PCR inhibitors on mRNA expression for human blood identification[J]. Legal Medicine, 2018, 32: 113-119. DOI:10.1016/j.legalmed.2018.04.002 |
| [15] |
Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction:Twenty-something years on[J]. Nature Protocols, 2006, 1(2): 581-585. DOI:10.1038/nprot.2006.83 |
| [16] |
宋洁云, 刘芳宏, 马军, 等. 酚/氯仿法和盐析法提取人类外周血基因组DNA方法的比较[J]. 中国实验诊断学, 2013, 17(5): 802-805. Song Jieyun, Liu Fanghong, Ma Jun, et al. Comparison of phenol/chloroform method and salting out method to extract human peripheral blood genomic DNA[J]. Chinese experimental diagnostics, 2013, 17(5): 802-805. DOI:10.3969/j.issn.1007-4287.2013.05.003 (in Chinese) |
| [17] |
Wright M H, Adelskov J, Greene A C. Bacterial DNA extraction using individual enzymes and phenol/chloroform separation[J]. Journal of Microbiology & Biology Education, 2017, 18(2): 1-3. |
| [18] |
Hayyan M, Looi C Y, Hayyan A, et al. In vitro and in vivo toxicity profiling of ammonium-based deep eutectic solvents[J]. PLoS One, 2015, 10(2): 1-18. |
| [19] |
Rahman M M, Elaissari A. Nucleic acid sample preparation for in vitro molecular diagnosis:From conventional techniques to biotechnology[J]. Drug Discovery Today, 2012, 17(21/22): 1199-1207. |
| [20] |
Chen H, Wu Y, Chen Z, et al. From sample to answer:A low-cost disposable cartridge for epidemic detection on site based on 3D printing technology[J]. Nanoscience and Nanotechnology Letters, 2016, 8(12): 1118-1126. DOI:10.1166/nnl.2016.2278 |
| [21] |
Gupta N. DNA extraction and polymerase chain reaction[J]. Journal of Cytology, 2019, 36(2): 116-117. DOI:10.4103/JOC.JOC_110_18 |
| [22] |
Hong S, Kim Y, Park J H. High-efficiency automated DNA extraction method for degraded old skeletal samples[J]. Forensic Science International:Genetics Supplement Series, 2017, 6: e365-e367. DOI:10.1016/j.fsigss.2017.09.108 |
| [23] |
Ayoib A, Hashim U, Gopinath S C B, et al. DNA extraction on bio-chip:History and preeminence over conventional and solid-phase extraction methods[J]. Applied Microbiology and Biotechnology, 2017, 101(22): 8077-8088. DOI:10.1007/s00253-017-8493-0 |
| [24] |
Pankhurst Q A, Jones S, Dobson J. Applications of magnetic nanoparticles in biomedicine:The story so far[J]. Journal of Physics D, 2016, 49(50): 1-3. |
| [25] |
Belanova A A, Gavalas N, Makarenko Y M, et al. Physicochemical properties of magnetic nanoparticles:Implications for biomedical applications in vitro and in vivo[J]. Oncology Research and Treatment, 2018, 41(3): 139-143. DOI:10.1159/000485020 |
| [26] |
Guo T, Lin M, Huang J, et al. The recent advances of magnetic nanoparticles in medicine[J]. Journal of Nanomaterials, 2018, 16: 1-8. |
| [27] |
Takabayashi S, Kotani S, Flores-Estrada J, et al. Boron-implanted silicon substrates for physical adsorption of DNA origami[J]. International Journal of Molecular Sciences, 2018, 19(9): 1-11. |
| [28] |
Urbaniak J, Janowski D, Jacewski B. Isolation of nucleic acids using silicon dioxide powder as a tool for environmental monitoring[J]. Environmental Monitoring and Assessment, 2019, 191(12): 1-7. |
| [29] |
Sano M, Kaji N, Wu Q, et al. Quantitative evaluation of dielectric breakdown of silicon micro- and nanofluidic devices for electrophoretic transport of a single DNA molecule[J]. Micromachines, 2018, 9(4): 1-12. |
| [30] |
Shuai H, Wu X, Huang K, et al. Ultrasensitive electrochemical biosensing platform based on spherical silicon dioxide/molybdenum selenide nanohybrids and triggered Hybridization Chain Reaction[J]. Biosensors and Bioelectronics, 2017, 94: 616-625. DOI:10.1016/j.bios.2017.03.058 |
| [31] |
An H, Jin B. Prospects of nanoparticle-DNA binding and its implications in medical biotechnology[J]. Biotechnology Advances, 2012, 30(6): 1721-1732. DOI:10.1016/j.biotechadv.2012.03.007 |
| [32] |
Kasyanenko N, Zhang Q, Bakulev V, et al. DNA binding with acetate bis(1, 10-phenanthroline)silver(I) monohydrate in a solution and metallization of formed structures[J]. Polymers, 2017, 9(6): 1-10. |
| [33] |
Zhang F, Sheng H, Wang S, et al. Screening DNA-targeted anticancer drug in vitro based on cancer cells DNA-templated silver nanoclusters[J]. Scientific Reports, 2019, 9(1): 1-8. DOI:10.1038/s41598-018-37186-2 |
| [34] |
Papagiannopoulos A, Mousdis G, Pispas S. Au nanoparticle-corona loaded polystyrene-b-quaternized poly(2-vinylpyridine) micelles and their interaction with DNA[J]. Macromolecular Chemistry and Physics, 2017, 218(3): 1-7. |
| [35] |
Wang L, Wang Y, Dong S L, et al. Nanocapsules of magnetic Au self-assembly for DNA migration and secondary self-assembly[J]. ACS Applied Materials & Interfaces, 2018, 10(6): 5348-5357. |
| [36] |
Sen R, Gahtory D, Carvalho R R, et al. Ultrathin covalently bound organic layers on mica:Formation of atomically flat biofunctionalizable surfaces[J]. Angewandte Chemie-International Edition, 2017, 56(15): 4130-4134. DOI:10.1002/anie.201701301 |
| [37] |
Yang J, Wang M, Cheng A, et al. A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection[J]. World Journal of Gastroenterology, 2008, 14(18): 2872-2876. DOI:10.3748/wjg.14.2872 |
| [38] |
Koyama H, Iwasa M, Tsuchimochi T, et al. Utility of Y-STR haplotype and mtDNA sequence in personal identification of human remains[J]. The American Journal of Forensic Medicine and Pathology, 2002, 23(2): 181-185. DOI:10.1097/00000433-200206000-00014 |
| [39] |
Zou Y, Mason M G, Wang Y, et al. Nucleic acid purification from plants, animals and microbes in under 30 seconds[J]. PLoS Biology, 2017. DOI:10.1016/j.ijmm.2017.11.004 |
| [40] |
Kim J H, Kim H J. Fast and simple method for screening of single-stranded DNA breaking photosensitizers using graphene oxide[J]. Nano Convergence, 2018, 5(1): 1-4. DOI:10.1186/s40580-017-0133-y |
| [41] |
Wu X, Mu F, Wang Y, et al. Graphene and graphene-based nanomaterials for DNA detection:A review[J]. Molecules, 2018, 23(8): 1-23. |
| [42] |
Close E D, Nwokeoji A O, Milton D, et al. Nucleic acid separations using superficially porous silica particles[J]. Journal of Chromatography A, 2016, 1440: 135-144. DOI:10.1016/j.chroma.2016.02.057 |
| [43] |
Quy D V, Hieu N M, Tra P T, et al. Synthesis of silica-coated magnetic nanoparticles and application in the detection of pathogenic viruses[J]. Journal of Nanomaterials, 2013, 200: 1-12. |
| [44] |
刘玲玲.赖氨酸修饰二氧化硅粒子的制备及其在DNA分离中的应用研究[D].长春: 吉林大学, 2016 Liu Lingling. Preparation of silica particles modified by lysine and its application in DNA separation[D]. Changchun: Ji Lin University, 2016(in Chinese) |
| [45] |
Gong D, Hui X, Guo Z, et al. The synthesis of PEI core@silica shell nanoparticles and its application for sensitive electrochemical detecting mi-RNA[J]. Talanta, 2019, 198: 534-541. DOI:10.1016/j.talanta.2019.02.013 |
| [46] |
Tiwari A P, Satvekar R K, Rohiwal S S, et al. Magneto-separation of genomic deoxyribose nucleic acid using pH responsive Fe3O4@silica@chitosan nanoparticles in biological samples[J]. RSC Advances, 2015, 5(11): 8463-8470. DOI:10.1039/C4RA15806G |
| [47] |
Jiang S, Zhuang J, Wang C, et al. Highly efficient adsorption of DNA on Fe3+-iminodiacetic acid modified silica particles[J]. Colloids and Surfaces A:Physicochemical and Engineering Aspects, 2012, 409: 143-148. DOI:10.1016/j.colsurfa.2012.05.051 |
| [48] |
Li T, Yi H, Liu Y, et al. One-step synthesis of DNA templated water-soluble Au-Ag bimetallic nanoclusters for ratiometric fluorescence detection of DNA[J]. Journal of Biomedical Nanotechnology, 2018, 14(1): 150-160. DOI:10.1166/jbn.2018.2491 |
| [49] |
Yang H. Highly sensitive electrochemical biosensor assembled by Au nanoparticle/MOF-5 composite electrode for DNA detection[J]. International Journal of Electrochemical Science, 2019, 5491-5507. DOI:10.20964/2019.06.49 |
| [50] |
Zhang M, Chen J, Pi X, et al. Construction and electrochemical property studies of DNA duplexes tethered to gold electrode via Au-C bond[J]. Electroanalysis, 2019, 31(3): 477-484. DOI:10.1002/elan.201800673 |
| [51] |
Sandstrom P, Boncheva M, Akerman B. Nonspecific and thiol-specific binding of DNA to gold nanoparticles[J]. Langmuir, 2003, 19(18): 7537-7543. DOI:10.1021/la034348u |
| [52] |
Thompson D G, Enright A, Faulds K, et al. Ultrasensitive DNA detection using oligonucleotide-silver nanoparticle conjugates[J]. Analytical Chemistry, 2008, 80(8): 2805-2810. DOI:10.1021/ac702403w |
| [53] |
Basu S M, Jana S, Pande S, et al. Interaction of DNA bases with silver nanoparticles:Assembly quantified through SPRS and SERS[J]. Journal of Colloid and Interface Science, 2008, 321(2): 288-293. DOI:10.1016/j.jcis.2008.02.015 |
| [54] |
Yang W, Shen C, Ji Q, et al. Food storage material silver nanoparticles interfere with DNA replication fidelity and bind with DNA[J]. Nanotechnology, 2009, 20(8): 1-8. |
| [55] |
Gu Q, Cheng C, Gonela R, et al. DNA nanowire fabrication[J]. Nanotechnology, 2006, 17(1): R14-R25. DOI:10.1088/0957-4484/17/1/R02 |
| [56] |
Rampini S, Li P, Lee G U. Micromagnet arrays enable precise manipulation of individual biological analyte-superparamagnetic bead complexes for separation and sensing[J]. Lab on a Chip, 2016, 16(19): 3645-3663. DOI:10.1039/C6LC00707D |
| [57] |
Ali Z, Wang J, Tang Y, et al. Simultaneous detection of multiple viruses based on chemiluminescence and magnetic separation[J]. Biomaterials Science, 2017, 5(1): 57-66. DOI:10.1039/C6BM00527F |
| [58] |
Wu Y, Chen H, Chen Z, et al. Robotic sample preparation system based on magnetic separation[J]. Journal of Nanoscience and Nanotechnology, 2016, 16(12): 12257-12262. DOI:10.1166/jnn.2016.12826 |
| [59] |
Veyret R, Delair T, Pichot C, et al. Amino-containing magnetic nanoemulsions:Elaboration and nucleic acid extraction[J]. Journal of Magnetism and Magnetic Materials, 2005, 295(2): 155-163. DOI:10.1016/j.jmmm.2005.01.008 |
| [60] |
高承刚, 李卫东, 杨晓蕾. DEAE-Sepharose FF纯化脊髓灰质炎病毒Sabin株Ⅰ型稳定性研究[J]. 西部医学, 2019, 31(6): 831-835. Gao Chenggang, Li Weidong, Yang Xiaolei. Study on the stability of DEAE-Sepharose FF purified poliovirus type 1 Sabin strain[J]. Medical Journal of West China, 2019, 31(6): 831-835. (in Chinese) |
| [61] |
Zheng M, Jagota A, Semke E D, et al. DNA-assisted dispersion and separation of carbon nanotubes[J]. Nature Materials, 2003, 2(5): 338-342. DOI:10.1038/nmat877 |
| [62] |
Zhao X, Striolo A, Cummings P T. C60 binds to and deforms nucleotides[J]. Biophysical Journal, 2005, 89(6): 3856-3862. DOI:10.1529/biophysj.105.064410 |
| [63] |
Hawkins T L, Mckernan K J, Jacotot L B, et al. DNA sequencing:A magnetic attraction to high-throughput genomics[J]. Science, 1997, 276(5320): 1887-1894. DOI:10.1126/science.276.5320.1887 |
| [64] |
Liu Z, Liu Y, Shen S, et al. Progress of recyclable magnetic particles for biomedical applications[J]. Journal of Materials Chemistry B, 2018, 6(3): 366-380. DOI:10.1039/C7TB02941A |
| [65] |
Bitar A, Vega-Chacon J, Lgourna Z, et al. Submicron silica shell-magnetic core preparation and characterization[J]. Colloids and Surfaces A-Physicochemical and Engineering Aspects, 2018, 537: 318-324. |
| [66] |
Tangchaikeeree T, Polpanich D, Elaissari A, et al. Magnetic particles for in vitro molecular diagnosis:From sample preparation to integration into microsystems[J]. Colloids and Surfaces B:Biointerfaces, 2017, 158: 1-8. DOI:10.1016/j.colsurfb.2017.06.024 |
| [67] |
Chen X, Mao Q, Liu J, et al. Isolation/separation of plasmid DNA using hemoglobin modified magnetic nanocomposites as solid-phase adsorbent[J]. Talanta, 2012, 100: 107-112. DOI:10.1016/j.talanta.2012.07.095 |
| [68] |
Płotka-Wasylka J, Szczepańska N, de la Guardia M, et al. Modern trends in solid phase extraction:New sorbent media[J]. TrAC Trends in Analytical Chemistry, 2016, 77: 23-43. DOI:10.1016/j.trac.2015.10.010 |
| [69] |
Chen Z, Wu Y, Kang M, et al. Research on automated nucleic acid extraction instrument based on magnetic nanoparticles separation[J]. Nanoscience and Nanotechnology Letters, 2018, 10(1): 60-68. DOI:10.1166/nnl.2018.2595 |
| [70] |
Stormer M, Kleesiek K, Dreier J. High-volume extraction of nucleic acids by magnetic bead technology for ultrasensitive detection of bacteria in blood components[J]. Clinical Chemistry, 2007, 53(1): 104-110. DOI:10.1373/clinchem.2006.070987 |
| [71] |
Berensmeier S. Magnetic particles for the separation and purification of nucleic acids[J]. Applied Microbiology and Biotechnology, 2006, 73(3): 495-504. DOI:10.1007/s00253-006-0675-0 |
| [72] |
Arriagada F J, Osseo-Asare K. Controlled hydrolysis of tetraethoxysilane in a nonionic water-in-oil microemulsion:A statistical model of silica nucleation[J]. Colloids and Surfaces A:Physicochemical and Engineering Aspects, 1999, 154(3): 311-326. |
| [73] |
Stober W, Fink A, Bohn E. Controlled growth of monogispherse silica sphrerrs in micro size range[J]. Journal of Colloid and Interface Science, 1968, 26(1): 62-69. DOI:10.1016/0021-9797(68)90272-5 |
| [74] |
Hui C, Shen C, Tian J, et al. Core-shell Fe3O4@SiO2 nanoparticles synthesized with well-dispersed hydrophilic Fe3O4 seeds[J]. Nanoscale, 2011, 3(2): 701-705. DOI:10.1039/C0NR00497A |
| [75] |
Yu X, Li P, Xiao C, et al. Synthesis of chain-like and core-shell spherical Fe3O4@SiO2 complex[J]. Advanced Science Letters, 2011, 4(1): 96-103. DOI:10.1166/asl.2011.1179 |
| [76] |
Yang P, Quan Z, Hou Z, et al. A magnetic, luminescent and mesoporous core-shell structured composite material as drug carrier[J]. Biomaterials, 2009, 30(27): 4786-4795. DOI:10.1016/j.biomaterials.2009.05.038 |
| [77] |
Abbas M, Abdel-Hamed M O, Chen J. Efficient one-pot sonochemical synthesis of thickness-controlled silica-coated superparamagnetic iron oxide (Fe3O4/SiO2) nanospheres[J]. Applied Physics A, 2017, 123(12): 1-6. |
| [78] |
Dehghan M, Motaharinejad A, Saadat M, et al. Novel approach to synthesizing polymer-functionalized Fe3O4/SiO2-NH2 via an ultrasound-assisted method for catalytic selective oxidation of alcohols to aldehydes and ketones in a DMSO/water mixture[J]. RSC Advances, 2015, 5(112): 92335-92343. DOI:10.1039/C5RA19093B |
| [79] |
Ma C, Li C, He N, et al. Preparation and characterization of monodisperse core-shell Fe3O4@SiO2 microspheres and its application for magnetic separation of nucleic acids from E. coli BL21[J]. Journal of Biomedical Nanotechnology, 2012, 8(6): 1000-1005. DOI:10.1166/jbn.2012.1454 |
| [80] |
Fan Q, Guan Y, Zhang Z, et al. A new method of synthesis well-dispersion and dense Fe3O4@SiO2 magnetic nanoparticles for DNA extraction[J]. Chemical Physics Letters, 2019, 715: 7-13. DOI:10.1016/j.cplett.2018.11.001 |
| [81] |
Quy D V, Hieu N M, Tra P T, et al. Synthesis of silica-coated magnetic nanoparticles and application in the detection of pathogenic viruses[J]. Journal of Nanomaterials, 2013, 2013: 1-7. |
| [82] |
Xu S, Song X, Guo J, et al. Composite microspheres for separation of plasmid DNA decorated with MNPS through in situ growth or interfacial immobilization followed by silica coating[J]. ACS Applied Materials & Interfaces, 2012, 4(9): 4764-4775. |
| [83] |
Deng Y, Qi D, Deng C, et al. Superparamagnetic high-magnetization microspheres with an Fe3O4@SiO2 core and perpendicularly aligned mesoporous SiO2 shell for removal of microcystins[J]. Journal of the American Chemical Society, 2008, 130(1): 28-29. DOI:10.1021/ja0777584 |
| [84] |
Liu Q, Li J, Liu H, et al. Rapid, cost-effective DNA quantification via a visually-detectable aggregation of superparamagnetic silica-magnetite nanoparticles[J]. Nano Research, 2014, 7(5): 755-764. DOI:10.1007/s12274-014-0436-9 |
| [85] |
He X, Huo H, Wang K, et al. Plasmid DNA isolation using amino-silica coated magnetic nanoparticles (ASMNPs)[J]. Talanta, 2007, 73(4): 764-769. DOI:10.1016/j.talanta.2007.04.056 |
| [86] |
Pham X H, Baek A, Kim T H, et al. Graphene oxide conjugated magnetic beads for RNA extraction[J]. Chemistry-an Asian Journal, 2017, 12(15): 1883-1888. DOI:10.1002/asia.201700554 |
| [87] |
Perçin I, Karakoç V, Akgöl S, et al. Poly(hydroxyethyl methacrylate) based magnetic nanoparticles for plasmid DNA purification from Escherichia coli lysate[J]. Materials Science and Engineering:C, 2012, 32(5): 1133-1140. DOI:10.1016/j.msec.2012.02.031 |
| [88] |
Zainabadi K, Adams M, Han Z Y, et al. A novel method for extracting nucleic acids from dried blood spots for ultrasensitive detection of low-density Plasmodium falciparum and Plasmodium vivax infections[J]. Malaria Journal, 2017, 16(1): 1-11. DOI:10.1186/s12936-016-1650-6 |
| [89] |
Tang Y, Zou J, Ma C, et al. Highly sensitive and rapid detection of pseudomonas aeruginosa based on magnetic enrichment and magnetic separation[J]. Theranostics, 2013, 3(2): 85-92. DOI:10.7150/thno.5588 |
| [90] |
Li P P, Mi R, Zhao R, et al. Quantitative real-time PCR with high-throughput automatable DNA preparation for molecular screening of Nosema SPP. in Antheraea pernyi[J]. Journal of Invertebrate Pathology, 2019, 164: 16-22. DOI:10.1016/j.jip.2019.04.003 |
| [91] |
Bai Y, Roncancio D, Suo Y, et al. A method based on amino-modified magnetic nanoparticles to extract DNA for PCR-based analysis[J]. Colloids and Surfaces B:Biointerfaces, 2019, 179: 87-93. DOI:10.1016/j.colsurfb.2019.03.005 |
| [92] |
Oblath E, Hampton Henley W, Alarie J P, et al. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva[J]. Lab on a Chip, 2013, 13(7): 1325-1332. DOI:10.1039/c3lc40961a |
| [93] |
汪浩, 朱元荣, 赵晓丽, 等. 常规金属离子对Fe3O4磁性纳米颗粒悬浮和沉降的影响[J]. 环境科学学报, 2017, 37(4): 1367-1373. Wang Hao, Zhu Yuanrong, Zhao Xiaoli, et al. Effect of conventional metal ions on suspension and deposition of Fe3O4 magnetic nanoparticles[J]. Journal of Environmental Science, 2017, 37(4): 1367-1373. (in Chinese) |
 2021, Vol. 38
2021, Vol. 38